Lesson: Nucleus
SYLLABUS
Nuclear Envelope- structure of nuclear pore complex;
chromatin;
molecular organization,
DNA packaging in eukaryotes,
euchromatin and heterochromatin,
nucleolus and ribosome structure (brief).
Glossary
Arginine: amino acid with the basic side/R group, so these are positively charged at pH 7.0
Condensins: it is a protein complex that results in metaphase chromosome condensation. These are members of a class of proteins, i.e., ‘structural maintenance of chromatin’(SMC).proteins
. Dinoflagellates: These are important components of communities of microrganisms that live on the waters surface of both fresh water as well as marine sources.
FISH: Fluorescent in situ hybridization. It is a technique in which fluorescent probes are used that bind to only those parts of the chromosomes in which sequence is complimentary to the sequence of the probe used. The technique is used to localize the presence or absence of specific DNA sequences on the chromosomes.
Lumen of ER: membranes of Endoplasmic reticulum divides the cytoplasm of the cell in two compartments, that is one enclosed within the membranes of endoplasmic reticulum which is called lumen or cisternal space, and the other outside i.e., cytosol.
Lysine: amino acid with the basic side/R group, so these are positively charged at pH 7.0
Nuclear localization signal: specific sequence of amino acids present on the N’terminal of the proteins which need to be transported to the nucleus.
Phosphorylation/dephosphorylation: addition/deletion of phosphate group to proteins or other organic compounds. Picogram:10 -12 gm
Protein kinase: It is a kinase enzyme that modifies other proteins by adding phosphate groups to them.
Spliceosomes: It is an RNA-protein complex that splices introns. From pre-mRNA and pastes the ends of the exons together.
Structural RNAs: these are non-coding RNAs.
Supercoiling: it refers to over or under-winding of DNA strand.
NUCLEUS
In cell biology, the nucleus (pl. nuclei; from Latin nucleus or nuculeus, meaning kernel) is a membrane-enclosed organelle found in eukaryotic cells. Eukaryotes usually have a single nucleus, but a few cell types have no nuclei, and a few others have many.
OR
DEFINITION
in eukaryotic cell, the most prominent organelle that contains the chromatin material, nucleolus and nuclear sap;surrounded by a double layered nuclear envelope is called nucleus.
FUNCTIONS OF NUCLEUS
It stores genetic information in the form of DNA
DNA is duplicated (replication) before it can be distributed equally into the daughter cells during the cell division.
mRNAs are synthesized (transcription) in the nucleus. These carry the genetic information from DNA in the nucleus to cytoplasm where the message is translated to proteins.
Ribosomes are synthesized in nucleolus, which are then transported to the cytoplasm.
Nuclear organization in eukaryotes
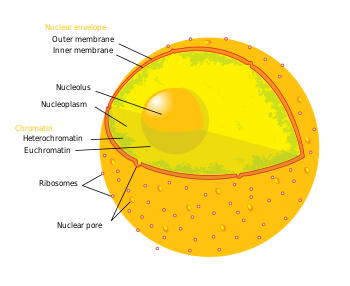
Nuclear envelope
Nuclear matrix and nuclear lamina
Nucleoplasm
Nucleolus
Chromatin
Nuclear envelope
protein complexes that cross the nuclear envelope, which is the double membrane surrounding
DEFINITION:
A nuclear membrane, also known as the nuclear envelope,nucleolemma or karyotheca, is the double lipid bilayer membrane which surrounds the genetic material and nucleolus in eukaryotic cells.
The nuclear membrane consists of two lipid bilayers—the inner nuclear membrane, and the outer nuclear membrane. The space between the membranes is called the perinuclear space, a region contiguous with the lumen (inside) of the endoplasmic reticulum. It is usually about 20–40 nm wide.
The nuclear membrane has many small holes called nuclear pores that allow material to move in and out of the nucleus.
In eukaryotes, nuclear compartment is separated from rest of the cell by means of nuclear envelope, which consists of two concentric unit membranes, i.e., outer nuclear membrane and inner nuclear membrane. J.Robertson (1960) had observed in the electron micrographs, that each membrane showed triple layered structure, i.e., dark–light-dark pattern. He proposed a model of membrane structure i.e., unit membrane model, in which lightly staining phospholipids layer is coated on its outer and inner surface by proteins in extended configuration. Each membrane is about 7-8 nm thick. These membranes enclose a space, called perinuclear space, which is about 20-40nm in width. Outer nuclear membrane is different from inner nuclear envelope in its chemical composition and function. The outer envelope is in direct contact with endoplasmic reticulum.
Membranes of both the organelles are similar in their structure and chemical composition. The thickness of both the membrane is similar and both provide the sites for attachment of ribosomes. However, no ribosomes are attached with the inner nuclear envelope. Inner nuclear envelope is closely associated with the chromatin.
The nuclear envelope is not continuous. A number of pores are present in it through which nucleoplasm is connected to cytoplasm. These pores are called nuclear pores. At the nuclear pore site, outer nuclear envelope is continuous with the inner nuclear envelope. Number of nuclear pores varies greatly with the cell type. There are about 10-20 pores per square micrometer in typical mammalian cells.
Nuclear pores are not simple pores but are rather complex in their structure. The structure is called nuclear pore complex (NPC). Diameter of the NPC is 120 nm. On the basis of studies of electron micrographs of NPC, structural models have been proposed. According to the models proposed, structure of NPC is thought to be like a basket, which fills the nuclear pore like a stopper. It consists of two rings, cytoplasmic ring (which is present towards cytoplasm) and nucleoplasmic ring (present towards nucleoplasm). Both the rings are connected to each other by means of spokes. Basket like structure, which is present towards nucleoplasm, is suspended by means of spokes. There is one central channel through which proteins and RNA are thought to pass through, while low molecular weight solute are believed to pass through the slots between the spoke. There are filaments present towards cytoplasmic side. These cytoplasmic filaments help movement of proteins through the pores. NPC is not a static structure, since many of its component proteins are replaced with new copies. NPC contains only about 30 different proteins, called nucleoporins. Among the nucleoporins, those which line the central channel of NPC, possess large number of phenylalanine-glycine repeats (FG-by their single letter code). This FG domains form a hydrophobic sieve which blocks the diffusion of larger macro-molecules (larger than about 40,000 daltons) between nucleus and cytoplasm.
Outer membrane
The outer nuclear membrane also shares a common border with endoplasmic reticulum. While it is physically linked, the outer nuclear membrane contains proteins found in far higher concentrations than the endoplasmic reticulum.Inner membrane
The inner nuclear membrane encloses the nucleoplasm, and is covered by the nuclear lamina, a mesh of intermediate filaments which stabilizes the nuclear membrane as well as being involved in chromatin function and entire expression. It is connected to the outer membrane by nuclear pores which penetrate the membranes. While the two membranes and the endoplasmic reticulum are linked, proteins embedded in the membranes tend to stay put rather than dispersing across the continuumNuclear pores
The nuclear membrane is punctured by thousands of nuclear pore complexes—large hollow proteins about 100 nm across, with an inner channel about 40 nm wide. They link the inner and outer nuclear membranes.Cell division
During the G2 phase of interphase, the nuclear membrane increases its surface area and doubles its number of nuclear pore complexes.In lower eukaryotes, such as yeast, which undergo closed mitosis, the nuclear membrane stays intact during cell division. The spindle fibers either form within the membrane, or penetrate it without tearing it apart.
In higher eukaryotes (animals), as well as plants, the nuclear membrane must break down during the prometaphase state of mitosis to allow the mitotic spindle fibers to access the chromosomes inside. The breakdown and reformation processes are not well understood.
Breakdown
In mammals, the nuclear membrane can break down within minutes, following a set of steps during the early stages of Mitosis.First, M-Cdk's phosphorylate nucleoporin polypeptides and they are selectively removed from the nuclear pore complexes. After that, the rest of the nuclear pore complexes break apart simultaneously. Biochemical evidence suggests that the nuclear pore complexes disassemble into stable pieces rather than disintegrating into small polypeptide fragments. M-Cdk's also phosphorylate elements of the nuclear lamina (the framework that supports the envelope) leading to the dis-assembly of the lamina and hence the envelope membranes into small vesicles.
Electron and fluorescence microscopy has given strong evidence that the nuclear membrane is absorbed by the endoplasmic reticulum—nuclear proteins not normally found in the endoplasmic reticulum show up during mitosis.
Reformation
Exactly how the nuclear membrane reforms during telophase of mitosis is debated. Two theories exist- Vesicle fusion—where vesicles of nuclear membrane fuse together to rebuild the nuclear membrane
- Reshaping of the endoplasmic reticulum—where the parts of the endoplasmic reticulum containing the absorbed nuclear membrane envelop the nuclear space, reforming a closed membrane.
Chromatin is a complex of DNA and proteins that forms chromosomes within the nucleus of eukaryotic cells. Nuclear DNA does not appear in free linear strands; it is highly condensed and wrapped around nuclear proteins in order to fit inside the nucleus.
Chromatin exists in two forms. One form, called euchromatin, is less condensed and can be transcribed. The second form, called heterochromatin, is highly condensed and is typically not transcribed.
Under the microscope in its extended form, chromatin looks like beads on a string. The beads are called nucleosomes. Each nucleosome is composed of DNA wrapped around eight proteins called histones. The nucleosomes are then wrapped into a 30 nm spiral called a solenoid, where additional histone proteins support the chromatin structure. During cell division, the structure of the chromatin and chromosomes are visible under a light microscope, and they change in shape as the DNA is duplicated and separated into two cells.
DNA packaging is an important process in living cells. Without it, a cell is not able to accommodate large amount of DNA that is stored inside. For example, a bacterial cell which ranges from 1 to 2um in length contains amount of DNA that is 400 times as big (Becker et al. 530). Eukaryotic cells face even bigger challenges. A typical human cell has enough “DNA to wrap around the cell more than 15,000 times” (531). Therefore, DNA packaging is crucial because it makes sure that those excessive DNA are able to fit nicely in a cell that is many times smaller.
The DNA in bacterial cells are either circular or linear. To accommodate the size of bacterial cell, supercoiled DNA are folded into loops with each loop resembles shape of bead-like packets containing small basic proteins that is analogous to histone found in Eukaryotes (533).
In eukaryotic cells, DNA packaging is more complicated because they contain amount of DNA that is much larger than that of bacterial cells. More proteins are therefore required for the process with histone being the most important one. This protein is consisted largely of positive amino acids like lysine and arginine which make the overall structure positive. Thus, histone interacts favorably with the negative phosphate groups from DNA. There are five main types of histone, H1, H2A, H2B, H3 and H4 (533). Two of each H2A, H2B, H3 and H4 joins to form an octamer wrapped around by DNA of 146 base pairs like a bead on a string. This bead, consisting of eight histone molecules and 146 DNA base pairs, is known as the nucleosome. Each nucleosome is connected by a DNA linker of 50 base pairs to form a fiber like structure called chromatin. H1 is believed to be found in these DNA linkers. Chromatin fibers can be further compacted to form higher order of structures called heterochromatin or euchromatin depending on the degree of packing. Ultimately, DNA packaging in eukaryotic cells can lead to the formation of chromosome which is only present during cell division or several other situations (533-535).
In eukaryotic cells, DNA packaging is not only in the nucleus but is also in mitochondria and chloroplast. The overall shape of their DNA resembles that of bacteria instead that of eukaryotes.
Chromatin is made of DNA and proteins (Histones). Chromatin is used to give structure to a chromosome.
Nucleosome consists of the acidic chromatin and the basic histone proteins.
Histone chaperones Histone chaperone guided folding pathways, assists in the folding and unfolding of the DNA around the histone.
Organization The tight coiling of DNA allows easier access to the DNA which makes sequencing faster.
Need for histone chaperones Nucleosomes can be assembled or disassembled and are done in stepwise function. Histone chaperones guide the pathway process, they control and regulate.
Structural forms of histone chaperones Since histone chaperones participate at each step of the nucleosome assembly processes, there are different chaperones needed for each different step.
Euchromatin
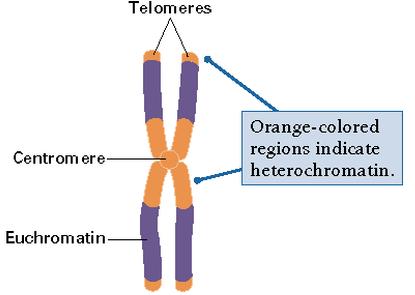
Euchromatin is a lightly packed form of chromatin (DNA, RNA and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin comprises the most active portion of the genome within the cell nucleus. 92% of the human genome is euchromatic.
The remainder is called heterochromatin
Chromatin exists in two forms. One form, called euchromatin, is less condensed and can be transcribed. The second form, called heterochromatin, is highly condensed and is typically not transcribed.
Under the microscope in its extended form, chromatin looks like beads on a string. The beads are called nucleosomes. Each nucleosome is composed of DNA wrapped around eight proteins called histones. The nucleosomes are then wrapped into a 30 nm spiral called a solenoid, where additional histone proteins support the chromatin structure. During cell division, the structure of the chromatin and chromosomes are visible under a light microscope, and they change in shape as the DNA is duplicated and separated into two cells.
DNA Packaging
The DNA in bacterial cells are either circular or linear. To accommodate the size of bacterial cell, supercoiled DNA are folded into loops with each loop resembles shape of bead-like packets containing small basic proteins that is analogous to histone found in Eukaryotes (533).
In eukaryotic cells, DNA packaging is more complicated because they contain amount of DNA that is much larger than that of bacterial cells. More proteins are therefore required for the process with histone being the most important one. This protein is consisted largely of positive amino acids like lysine and arginine which make the overall structure positive. Thus, histone interacts favorably with the negative phosphate groups from DNA. There are five main types of histone, H1, H2A, H2B, H3 and H4 (533). Two of each H2A, H2B, H3 and H4 joins to form an octamer wrapped around by DNA of 146 base pairs like a bead on a string. This bead, consisting of eight histone molecules and 146 DNA base pairs, is known as the nucleosome. Each nucleosome is connected by a DNA linker of 50 base pairs to form a fiber like structure called chromatin. H1 is believed to be found in these DNA linkers. Chromatin fibers can be further compacted to form higher order of structures called heterochromatin or euchromatin depending on the degree of packing. Ultimately, DNA packaging in eukaryotic cells can lead to the formation of chromosome which is only present during cell division or several other situations (533-535).
In eukaryotic cells, DNA packaging is not only in the nucleus but is also in mitochondria and chloroplast. The overall shape of their DNA resembles that of bacteria instead that of eukaryotes.
Histone chaperones and the nucleosome assembly processes
Histones are proteins that allow DNA to be tightly packaged into units called nucleosomes. The DNA wraps itself around the histones.Chromatin is made of DNA and proteins (Histones). Chromatin is used to give structure to a chromosome.
Nucleosome consists of the acidic chromatin and the basic histone proteins.
Histone chaperones Histone chaperone guided folding pathways, assists in the folding and unfolding of the DNA around the histone.
Organization The tight coiling of DNA allows easier access to the DNA which makes sequencing faster.
Need for histone chaperones Nucleosomes can be assembled or disassembled and are done in stepwise function. Histone chaperones guide the pathway process, they control and regulate.
Structural forms of histone chaperones Since histone chaperones participate at each step of the nucleosome assembly processes, there are different chaperones needed for each different step.
Euchromatin

Euchromatin is a lightly packed form of chromatin (DNA, RNA and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin comprises the most active portion of the genome within the cell nucleus. 92% of the human genome is euchromatic.
The remainder is called heterochromatin
Structure
The structure of euchromatin is reminiscent of an unfolded set of beads along a string, wherein those beads represent nucleosomes. Nucleosomes consist of eight proteins known as histones, with approximately 147 base pairs of DNA wound around them; in euchromatin, this wrapping is loose so that the raw DNA may be accessed. Each core histone possesses a `tail' structure, which can vary in several ways; it is thought that these variations act as "master control switches," which determine the overall arrangement of the chromatin. In particular, it is believed that the presence of methylated lysine 4 on the histone tails acts as a general marker for euchromatinAppearance
In general, euchromatin appears as light-colored bands when stained in G banding[citation needed] and observed under an optical microscope, in contrast to heterochromatin, which stains darkly. This lighter staining is due to the less compact structure of euchromatin. The basic structure of euchromatin is an elongated, open, 10 nm microfibril, as noted by electron microscopy. In prokaryotes, euchromatin is the only form of chromatin present; this indicates that the heterochromatin structure evolved later along with the nucleus, possibly as a mechanism to handle increasing genome size.Function
Euchromatin participates in the active transcription of DNA to mRNA products. The unfolded structure allows gene regulatory proteins and RNA polymerase complexes to bind to the DNA sequence, which can then initiate the transcription process. Not all euchromatin is necessarily transcribed, but in general that which is not is transformed into heterochromatin to protect the genes while they are not in use. There is therefore a direct link to how actively productive a cell is and the amount of euchromatin that can be found in its nucleus. It is thought that the cell uses transformation from euchromatin into heterochromatin as a method of controlling gene expression and replication, since such processes behave differently on densely compacted chromatin, known as the `accessibility hypothesis'. One example of constitutive euchromatin that is 'always turned on' is housekeeping genes, which code for the proteins needed for basic functions of cell survival.Constitutive heterochromatin
Constitutive heterochromatin domains are regions of DNA found throughout the chromosomes of eukaryotes. The majority of constitutive heterochromatin is found at the pericentromeric regions of chromosomes, but is also found at the telomeres and throughout the chromosomes. In humans there is significantly more constitutive heterochromatin found on chromosomes 1, 9, 16, 19 and on the Y chromosome. Constitutive heterochromatin is composed mainly of high copy number tandem repeats known as satellite repeats, minisatellite and microsatellite repeats, and transposon repeats. In humans these regions account for about 200Mb or 6.5% of the total human genome, but their repeat composition has made it difficult to sequence so only small regions have been sequenced. The Y chromosome is an exception in that it has been sequenced because its repeats do not cause difficulties in finding overlapping regions that are needed for sequencing.
Visualization of constitutive heterochromatin is possible by using the C-banding technique.
The regions that stain darker are regions of constitutive heterochromatin. The constitutive heterochromatin stains darker because of the highly condensed nature of the DNA.
Constitutive heterochromatin is not to be confused with faculatitive heterochromatin which is less condensed, less stable, does not stain when using the C-banding technique, and is much less polymorphic.
Constitutive heterochromatin was thought to be relatively devoid of genes, but researchers have found more than 450 genes in the heterochromatic DNA of Drosophila melanogaster. These regions are highly condensed and epigenetically modified to prevent transcription. In order for the genes to be transcribed they must have a mechanism to overcome the silencing that occurs in the rest of the heterochromatin. There are many proposed models for how the genes in these regions are expressed, including the insulation, denial, integration, exploitation, and TE restraining models.
In most cases when genes are placed near a region of constitutive heterochromatin their transcription is silenced, this effect is known as position-effect variegation and can lead to a mosaic phenotype
Visualization of constitutive heterochromatin is possible by using the C-banding technique.
The regions that stain darker are regions of constitutive heterochromatin. The constitutive heterochromatin stains darker because of the highly condensed nature of the DNA.
Constitutive heterochromatin is not to be confused with faculatitive heterochromatin which is less condensed, less stable, does not stain when using the C-banding technique, and is much less polymorphic.
Function
Constitutive heterochromatin is found more commonly in the periphery of the nucleus attached to the nuclear membrane. This concentrates the euchromatic DNA in the center of the nucleus where it can be actively transcribed. During mitosis it is believed that constitutive heterochromatin is necessary for proper segregation of sister chromatids and centromere function. The repeat sequences found at the pericentromeres is not conserved throughout many species and depends more on the epigenetic modifications for regulation while telomeres show more conserved sequences.Constitutive heterochromatin was thought to be relatively devoid of genes, but researchers have found more than 450 genes in the heterochromatic DNA of Drosophila melanogaster. These regions are highly condensed and epigenetically modified to prevent transcription. In order for the genes to be transcribed they must have a mechanism to overcome the silencing that occurs in the rest of the heterochromatin. There are many proposed models for how the genes in these regions are expressed, including the insulation, denial, integration, exploitation, and TE restraining models.
In most cases when genes are placed near a region of constitutive heterochromatin their transcription is silenced, this effect is known as position-effect variegation and can lead to a mosaic phenotype
Nucleolous

The nucleolus is the largest structure in the nucleus of eukaryotic cells, where it primarily serves as the site of ribosome synthesis and assembly. Nucleoli also have other important functions like assembly of signal recognition particles and playing a role in the cell's response to stress. Nucleoli are made of proteins and RNA and form around specific chromosomal regions. Malfunction of nucleoli can be the cause of several human diseases.
Structure
Three major components of the nucleolus are recognized: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). The DFC consists of newly transcribed rRNA bound to ribosomal proteins, while the GC contains RNA bound to ribosomal proteins that are being assembled into immature ribosomes.However, it has been proposed that this particular organization is only observed in higher eukaryotes and that it evolved from a bipartite organization with the transition from anamniotes to amniotes. Reflecting the substantial increase in the DNA intergenic region, an original fibrillar component would have separated into the FC and the DFC.
Another structure identified within many nucleoli (particularly in plants) is a clear area in the center of the structure referred to as a nucleolar vacuole. Nucleoli of various plant species have been shown to have very high concentrations of iron in contrast to human and animal cell nucleoli.
The nucleolus ultrastructure can be visualized through an electron microscope, while the organization and dynamics can be studied through fluorescent protein tagging and fluorescent recovery after photobleaching (FRAP). Antibodies against the PAF49 protein can also be used as a marker for the nucleolus in immunofluorescence experiments.
Function of Nucleolus
| The nucleus of many eukaryotic cells contains a structure called a nucleolus. As the nucleus is the "brain" of the cell, the nucleolus could loosely be thought of as the brain of the nucleus. The nucleolus takes up around 25% of the volume of the nucleus. This structure is made up of proteins and ribonucleic acids (RNA). Its main function is to rewrite ribosomal RNA (rRNA) and combine it with proteins. This results in the formation of incomplete ribosomes. There is an uninterrupted chain between the nucleoplasm and the interior parts of the nucleolus, whichoccurs through a system of nucleolarpassages. These passages allow macromolecules with a molecular weight up to 2,000 kDato be easily circulated throughout the nucleolus. Because of its close relationship to the chromosomal matter of the cell and its important role in producing ribosomes, the nucleolus is thought to be the cause of a variety of different human diseases. |
The ribosome is a complex molecular machine found within all living cells, that serves as the site of biological protein synthesis (translation). Ribosomes link amino acids together in the order specified by messenger RNA (mRNA) molecules. Ribosomes consist of two major components: the small ribosomal subunit, which reads the RNA, and the large subunit, which joins amino acids to form a polypeptide chain. Each subunit is composed of one or more ribosomal RNA (rRNA) molecules and a variety of proteins. The ribosomes and associated molecules are also known as the translational apparatus

Figure : Atomic structure of the 30S Subunit from Thermus thermophilus.[8] Proteins are shown in blue and the single RNA chain in orange
Ribosomes are the workplaces of protein biosynthesis, the process of translating mRNA into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon (3 nucleotides) of the mRNA, pairing it with the appropriate amino acid provided by an aminoacyl-tRNA. Aminoacyl-tRNA contains a complementary anticodon on one end and the appropriate amino acid on the other. For fast and accurate recognition of the appropriate tRNA, the ribosome utilizes large conformational changes (conformational proofreading) . The small ribosomal subunit, typically bound to an aminoacyl-tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The ribosome contains three RNA binding sites, designated A, P and E. The A site binds an aminoacyl-tRNA; the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome. Protein synthesis begins at a start codon AUG near the 5' end of the mRNA. mRNA binds to the P site of the ribosome first. The ribosome is able to identify the start codon by use of the Shine-Dalgarno sequence of the mRNA in prokaryotes and Kozak box in eukaryotes.
Although catalysis of the peptide bond involves the C2 hydroxyl of RNA's P-site adenosine in a proton shuttle mechanism, other steps in protein synthesis (such as translocation) are caused by changes in protein conformations. Since their catalytic core is made of RNA, ribosomes are classified as "ribozymes,"[28] and it is thought that they might be remnants of the RNA world.[29]
Addition of translation-independent amino acids
Presence of a ribosome quality control protein Rqc2 is associated with mRNA-independent protein elongation.[30][31] This elongation is a result of ribosomal addition (via tRNAs brought by Rqc2) of CAT tails: ribosomes extend the C-terminus of a stalled protein with random, translation-independent sequences of alanines and threonines.
VIKAS BHATI








No comments:
Post a Comment